Tandem Mass Spectrometry and Peptide Fragmentation
1. Tandem mass spectrometry (MS/MS) can be achieved in three steps through two modes, tandem in time and in space.
- What are those three steps?
mass filter
fragmentation
detect & mass analysis - Which single mass analyzer(s) alone can achieve MS/MS?
ion trap and FT-ICR - There are three mass analyzers in the new Thermo Fisher Obtriap Fusion. What are these three analyzers?
Quadrupole Mass Filter
Ultra-High-Field Orbitrap Mass Analyzer
Dual-Pressure Linear Ion Trap
2. One chemical reagent iodoacetamide is commonly used during the generation of peptides. What is the purpose of the reagent?
For alkylation of cysteines in proteomics
3. What is the specificity of trypsin for protein digestion? Why is trypsin is so commonly used for proteomics?
Lys and Arg
- It is cheap
- Its specificity (Lys and Arg) mean that, on average, it will produce lots of peptides from a protein that are in the ideal mass range for MS analysis (~500-2500 Da)
- The specificity guarantees that at least one basic residue that can carry a positive charge will be present (for ESI it means that doubly charged peptides will be preferred which enhances tandem MS utility)
- It is highly specific (very little non-tryptic cleavage) and active under a wide variety of conditions, including fairly strong denaturing conditions
- Works equally well for solution and in-gel digest formats
4. Can you draw a generic structure of tripeptide and indicate the formation of a,b,c, and x,y,z ions?
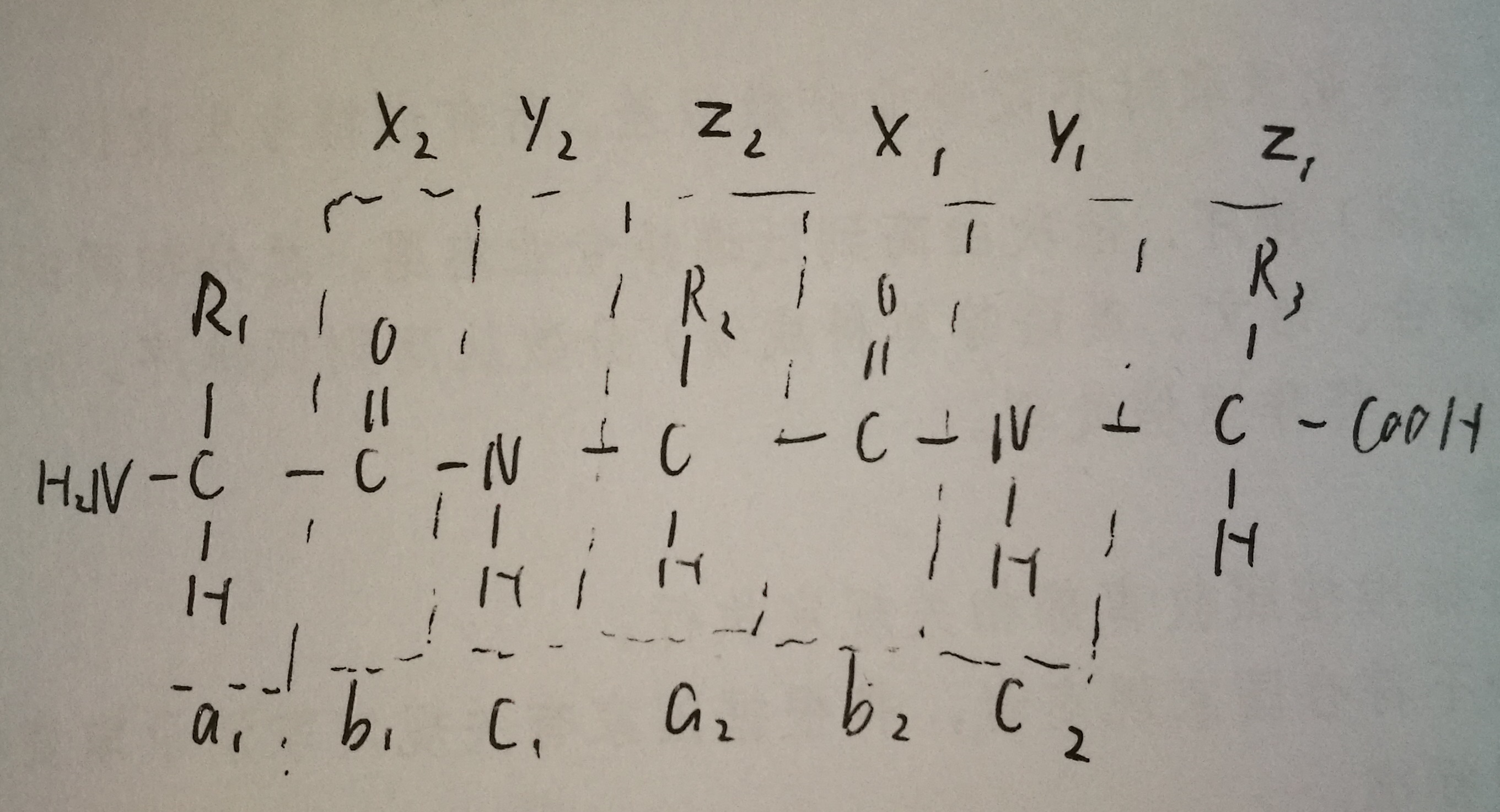
5. Give one major difference between CID and ETD and one specific application of ETD.
| Differences | CID | ETD |
|---|---|---|
| Charge | works best on doubly charged peptides | requires +3 charge or higher |
| Location | results in primarily amide bond cleavage generating y and b ions | results in N-Cα bond cleave generating c and z ions |
| Energy | relatively higher energy process causing fragmentation of labile modifications (e.g. phosphorylation, glycosylation) | very low-energy process that maintains modifications |
analyze protein modification
Protein identification by MS
6. What is the PMF? Which instrument you shall choose for PMF, TOF or ion trap? Please state your reason.
Peptide mass fingerprinting (PMF) (also known as protein fingerprinting) is an analytical technique for protein identification in which the unknown protein of interest is first cleaved into smaller peptides, whose absolute masses can be accurately measured with a mass spectrometer such as MALDI-TOF or ESI-TOF.
I choose TOF because of its high mass accuracy as well as speed and high throughput. The procedure is entirely dependent on accurate measured masses of proteolytic peptides.
7. Determine as much of the amino acid sequence as possible of the tryptic peptide represented by the MALDI MS/MS spectrum shown below obtained on a TOF-TOF instrument. The parent ion was 1606.71+1. Label all y and b ions that were found and the amino acids that correspond to each gap between y and b ions. Make sure you list your final sequence clearly as well.
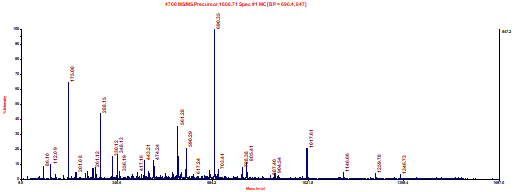
L/I F S L/I E L/I V D E S G E L/I R
8. Determine the amino acid sequence of the tryptic peptide represented by the ESI MS/MS spectrum shown below obtained on a Q-TOF instrument. Label all y and b ions found in the spectrum and label the gaps between peaks with the corresponding amino acid residue. The parent ion was 460.3+2. Make sure to provide your final sequence as well.
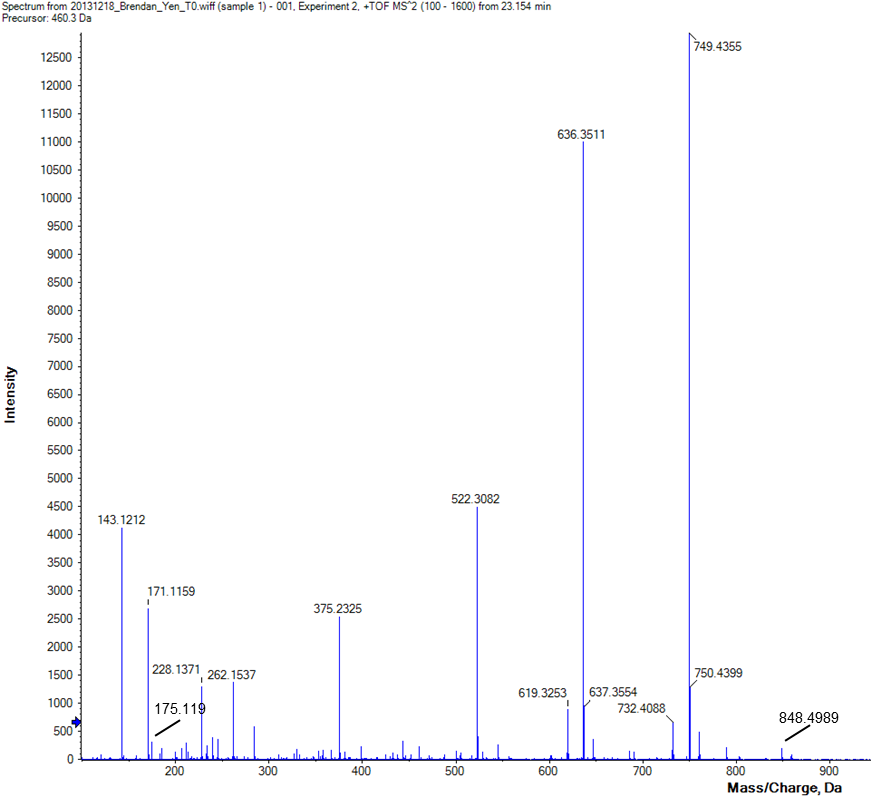
A V L/I N F L/I S R